Fast DNA amplification with miniPCR
Robust PCR performance in 24 minutes
Being able to do high-speed PCR, especially in time constrained settings such as STEM education environments, is a key step to make molecular biology accessible and practicable. Below we share how the miniPCR™ machine can be used to deliver robust and reproducible PCR results in 24 minutes or less.
Polymerase chain reaction (PCR) is the process of amplifying targeted copies of DNA by the billions. From the sound of it, one would think that this should take a long time–just thinking about the amount of DNA produced. In time-constrained settings, such as the classroom, PCR often seems unfathomable, but below we show that it can be done. Fast.
We set out to challenge the idea that PCR needs to take one hour or more by first identifying an adequate low-complexity target. Because we wanted this example to be realistic, we aimed to use tiny (low picogram) amounts of plasmid DNA cloned in our own lab. And because we wanted it to be directly classroom-relevant, we settled for pYUS5, a recombinant DNA plasmid carrying endogenous plant tubulin sequences, often used as a positive control in plant transformation experiments.
The DNA template input was 4 picograms of pYUS5. Samples were incubated with 5X EZ PCR Master Mix (PCR buffer with Mg2+, Taq polymerase, and dNTPs) and the miniPCR GMO Lab primers (0.1 µM of each primer). The target amplicon is 195 base pairs long. The PCR cycling parameters were:
- 94˚C initial denaturation, 10 sec
- 94˚C denaturation, 4 sec
- 65˚C annealing, 4 sec
- 72˚C extension, 1 sec
- Repeat x 25 cycles
- 72˚C Final extension, 10 sec

The PCR run took 24 minutes and 23 seconds in a miniPCR mini8 thermal cycler, including the initial lid heating step. Now was the time to see if this fast PCR run actually generated enough DNA to be seen in an agarose gel. After 25 cycles, 12 µl of each sample and controls were loaded into a 2.4% agarose gel using blueGel™ electrophoresis system, run for 25 minutes, and visualized using GelGreen™ DNA stain. Results are shown below:
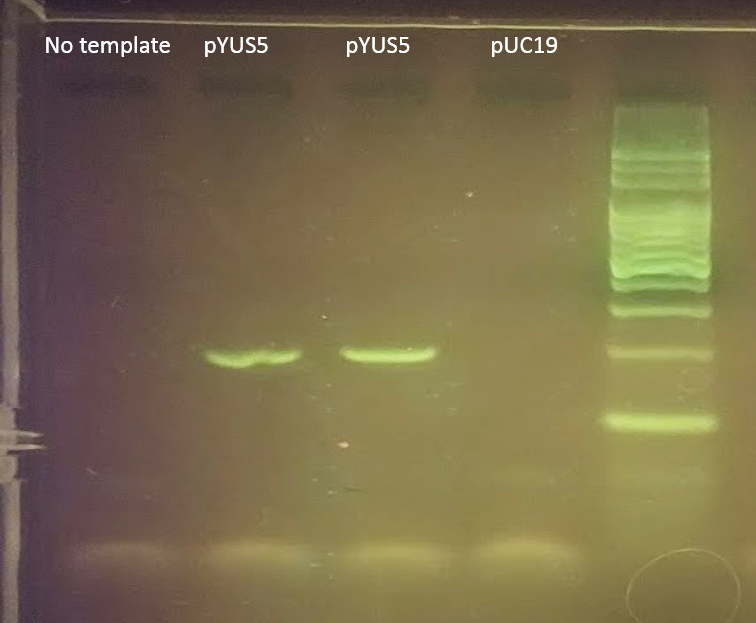
This 24-minute 23-second run yielded robust and reproducible results. Negative controls (no DNA input and a non-target plasmid) yielded no amplification bands, as expected. We hypothesize that further reductions in run time may be possible by further reducing step times (e.g. eliminate initial denaturation and final extension) or by reducing the number of cycles. Additional testing will likely confirm these predictions. Through these trials we can conclude that in a time-constrained environment, PCR can be completed in sub-25 minute time frames using standard reagents available in an educational kit. Several other plasmids and primer combinations could have been used to yield similar results in a classroom setting.
It does not escape our notice that many commercially available PCR machines should also be able to run a successful PCR within a similar time frame, assuming adequate ramp times and comparable heat transfer performance to miniPCR’s small thermal mass sample block. The miniPCR™ machine was developed to make the process of PCR more accessible and practicable. Being able to do high-speed PCR, especially in time constrained settings such as STEM education environments, is a great step in that direction.
If you have questions about implementing fast PCR, please contact us.