Demystifying another PCR myth: Ultra-pure water
Myth: PCR always needs to be conducted in ultrapure PCR-grade water
Truth: Many PCR experiments produce robust DNA amplification in tap water!
After securing lab space and obtaining lab equipment, the next barrier that independent or under-resourced scientists face is the high cost of molecular biology reagents and consumables. This naturally includes plastic ware, enzymes, synthetic DNA oligos, and buffers. Interestingly, there’s also a perceived generalized need for special water for PCR experiments. This comes in the shape of “ultra-pure”, “molecular grade” and “PCR-grade” water which can cost over $5,000 per liter[1]!
“PCR water“ is typically certified to be free of nucleases, so that DNA product do not get degraded, and free of nucleic acid contamination, so that the PCR does not yield false positives. In addition, “PCR water” is ion-free (i.e. distilled H2O) as a way to minimize interference with the activity of the DNA polymerase. Purchasing ultra-pure “PCR water” might be justified for some experiments, for example to avoid unwanted contamination when attempting to detect naturally-occurring nucleic acids that are ubiquitously expressed, such as those coding for 16S ribosomal RNAs. But we hypothesized that this precaution might be excessive for a vast number of other PCR applications, such as when amplifying uncommon DNA sequences from plasmids or other abundant sources. For example, when trying to generate fluorescent bacteria or bacteria that smell like banana the DNA sequences amplified are so specific and rare in nature that there is little risk of contamination with any nucleic acids that may be present in the water.
We wondered whether the use of regular “open source” (read: tap) water would impact PCR outcomes. Back in our academic research labs, this experiment wouldn’t even have crossed our minds (at least we would not have told anyone about it). Equipped with miniPCR, we set out to conduct an assessment of the feasibility of using tap water as the main solvent in PCR.
We started by identifying experimental “open source” water (i.e. tap water). Living in Somerville, Massachusetts, our communities are supplied by the Massachusetts Water Resources Authority (MWRA). The MWRA obtains its water supply from the Quabbin Reservoir, the Wachusett Reservoir, and the Ware River which have a combined capacity of approximately 477 billion gallons[2]. According to the Boston Sewer and Water Commission, “the typical customer pays just over a penny per gallon” of water supplied by the MWRA. The water for the neighboring city of Cambridge comes from a different source, the Hobbs Brook and Stony Brook reservoirs. In addition to Somerville and Cambridge tap waters, we splurged and also tested regular bottled water (Crystal Geyser), and distilled water (Market Basket, $0.99 per gallon) both purchased at our local grocery store. Our “PCR water” control was double-distilled, Milli-Q filtered water, kindly donated by a local research lab (even though Milli-Q filtered water is not considered by some as clean as commercial PCR-grade water, many labs use Milli-Q water for their everyday PCR needs.)
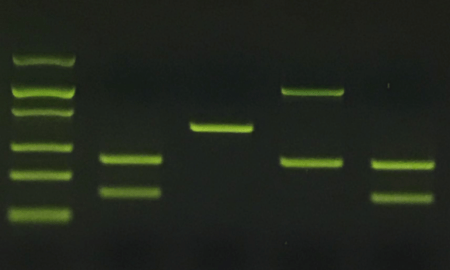
We used the different waters as solvents in PCR experiments targeting a 380-bp region of plasmid DNA (pMAL c5-e, New England Biolabs) using 30 cycles of amplification. In every case, test water accounted for 50% of the volume of the PCR mix (25 of 50 microliters). To challenge the robustness of PCR and reveal potential differences between the waters, we used stringent PCR conditions (such as nanogram template DNA, sub-micromolar primer concentrations, and denaturing, annealing and extension times of ten seconds for each cycle.) The Taq DNA polymerase was EZ PCR Master Mix, and the final MgCl2 concentration 1.5 mM.

The result, likely surprising to many, is that regular tap water yielded PCR results. Samples amplified in Milli-Q filtered water showed similar amplification to all other waters tested, at least under our experimental conditions (380bp amplicon from a plasmid DNA template.) It is likely that more restrictive PCR conditions or the use of a more complex DNA template could result in more variable amplification across samples.
We’re looking forward to assessing the repeatability of our results. Because the public water supply is under rigorous monitoring of its microbiological and mineral composition, we expect this result to be consistent over time. An intriguing follow up assessment would be to try amplification of naturally occurring microbial templates, such as ribosomal DNA sequences, and assess the possible occurrence of false positives.
Several PCR applications such as Reverse-Transcriptase PCR where the starting material is RNA, diagnostic PCR used in microbiology for the specific detection of microorganisms, semi-quantitative or “detection limit” PCR, will of course require the use of sterile and clean sources of water (RNAse and DNAse free, DNA free, autoclaved and UV irradiated, etc.). But we can confidently recommend the use of widely available, “open source” tap water or inexpensive dH2O for a large number of non-diagnostic PCR applications. In many of the contamination-sensitive cases, the addition of “blank” samples omitting the DNA template can provide the right level of negative control to account for potential contaminants in the water.
References
[1]
https://www.lifetechnologies.com/order/catalog/product/AM9935 $89.75/10ml = $8,975/l
https://www.mobio.com/pcr-water/ $67/10ml = $6,700/l
https://lifescience.roche.com/shop/products/water-pcr-grade $137/25ml = $5,480/l
[2]
https://www.bwsc.org/ABOUT_BWSC/systems/water/PresentDay_water.asp
https://en.wikipedia.org/wiki/Boston_Water_and_Sewer_Commission
About the Authors and about miniPCR bio
Ezequiel Alvarez-Saavedra, PhD, co-founded miniPCR to help make science simpler and more accessible to more people. Zeke has conducted biomedical research alongside two Nobel laureates and is the author of multiple publications in top peer-reviewed scientific journals. His work has been cited thousands of times and profiled in national and international media such as The New York Times, National Public Radio and the BBC. He is an inventor of patented gene-detection technologies. Zeke studied Biology at the University of Buenos Aires and Stanford University, and obtained his PhD in Biology from MIT. zeke@minipcr.com
Sebastian Kraves, PhD, co-founded miniPCR to help bring science to more people in more places. He previously worked on making biomedical technology accessible with the world’s leading philanthropies, corporations, and multilateral organizations. Sebastian has published widely cited work on neural circuits, optogenetics, and the molecular regulation of circadian behavior. He studied Economics and Biology at the University of Buenos Aires, obtained his Doctorate in Neurobiology from Harvard Medical School, and was a postdoctoral fellow at Harvard University. seb@minipcr.com
miniPCR and the miniPCR logo are trademarks of Amplyus LLC