Learning Labs on Antibiotic Resistance
The spread of antibiotic resistance in bacteria is one of the great public health challenges of our time. We now offer three engaging Learning Labs you can use to teach your students about this pressing issue in an interactive and hands-on way. Which approach is right for you?
- Agricultural Monitoring Lab: Monitoring Resistant Organisms in the Environment:
 Developed in collaboration with the Prevalence of Antibiotic Resistance in the Environment (PARE) project at Tufts University, this lab presents students with a case study: dangerous carbapenem resistant bacteria have been identified as spreading in some farms. Your students must test the environmental DNA (eDNA) from two farms to see if the animals there are at risk. This approach offers high quality curriculum in a format that is easy to implement. Importantly, because DNA samples are provided and no DNA extraction is needed, results tend to be incredibly robust, even for those new to the molecular lab.
Developed in collaboration with the Prevalence of Antibiotic Resistance in the Environment (PARE) project at Tufts University, this lab presents students with a case study: dangerous carbapenem resistant bacteria have been identified as spreading in some farms. Your students must test the environmental DNA (eDNA) from two farms to see if the animals there are at risk. This approach offers high quality curriculum in a format that is easy to implement. Importantly, because DNA samples are provided and no DNA extraction is needed, results tend to be incredibly robust, even for those new to the molecular lab. - eDNA Project: Sampling Soil for Antibiotic Resistance: This open-ended lab allows students to test soils directly and in the process participate in the PARE project’s international antibiotic resistance monitoring program. Students can collect soil based on their own hypotheses of where antibiotic resistance may or may not be common. They then extract total eDNA and use PCR to test for two common tetracycline resistance genes. Finally, students upload their results to the PARE database, helping scientists gain an understanding of where antibiotic resistance is most likely to be found. As an advanced procedure with unknown outcomes, this lab works great for advanced lab classes, independent research projects, and course-based undergraduate research experiences (CUREs).
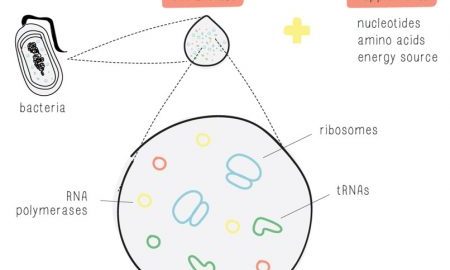
- BioBits®: Antibiotic Resistance: This hands-on activity reveals molecular targets of antibiotics and antibiotic resistance genes. Using a cell-free system with fluorescent readouts, students will study the effect of two antibiotics on bacterial protein synthesis. Students will also examine how resistance genes act in an antibiotic-specific manner. This laboratory exercise promotes a detailed understanding of antibiotic mechanisms and bacterial resistance.
Why choose just one? When used together, these labs provide an excellent scaffolded approach. First, use BioBits: Antibiotic Resistance to explore the mechanisms of antibiotic resistance. Then, use the Agricultural Monitoring Lab to learn about antibiotic resistance as an environmental problem, in a format with reliable results. Finally, students apply what they’ve learned in an authentic and meaningful way using the eDNA Project.
| Agricultural Monitoring Lab | eDNA Project | BioBits®: Antibiotic Resistance | |
|---|---|---|---|
| Approach | Case study format | Authentic soil sampling | Explore antibiotic resistance mechanisms in a cell-free system |
| Source of DNA samples | Prepared DNA samples guarantee robust, repeatable results | Environmental DNA extracted from student-gathered soil provides an authentic research experience with unknown outcomes | Prepared DNA samples guarantee robust, repeatable results |
| Best use | Traditional lab classes in general high school, advanced high school, or college | Advanced high school and college classes with molecular biology experience. Also suited for advanced lab classes, CUREs, and independent research | Traditional lab classes in general high school, advanced high school, or college |
| Techniques |
|
|
|
Related resources: