DNA Barcoding for Beginners
DNA barcoding is a specific type of DNA analysis aimed at identifying different species. This can be used by citizen scientists who want to identify mushrooms from their local forests, by field researchers measuring biodiversity in a remote jungle, or by food safety experts monitoring for harmful bacterial outbreaks. In this blog post, we’ll break down the basics of DNA barcoding and share some Learning Labs™ where you can identify species in your own environment!
What is a DNA barcode?
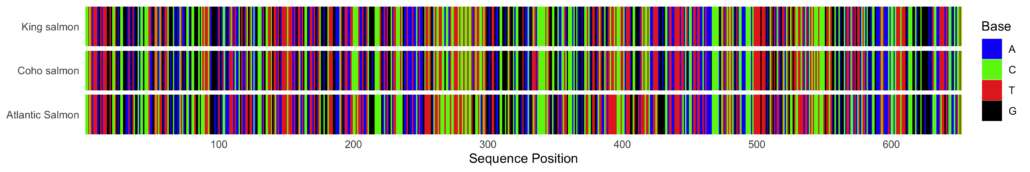
In the same way that a barcode on an item at the grocery store serves as a unique identifier linked to product information, DNA barcodes serve as unique identifiers for species. All living organisms have shared DNA sequences, so when looking for barcodes scientists focus on regions of DNA that are the same across individuals of the same species but diverge in members of other species. When displayed graphically, a DNA barcode resembles the lines we associate with a product barcode and can help you identify where organisms have shared sequences and regions of variability in their DNA.
How are DNA barcodes selected?
Scientists select short regions of DNA that are generally conserved within large groups of organisms, but are variable across species within that group. For example, the ITS region is a DNA barcode commonly used for fungi, while a region of the COI gene serves as a DNA barcode for most animals. The sequence of DNA bases in these regions are largely similar, but they include small variations unique to each species. Even closely related species that may look very similar to our eyes have enough variation in these DNA barcode regions that we can distinguish them using DNA analysis.

What steps are used in DNA barcoding?
- DNA extraction: DNA is removed from the cells of the organism you wish to identify. It is important that your samples be homogenous and not represent a mixture of species.
- PCR amplification of the DNA barcode: Polymerase chain reaction is used to copy a target region of DNA specific to the type of organism (i.e. fungi, bacteria, etc.). Primers that are specific to the regions being amplified are among the reagents included in this reaction. You may also choose to confirm that the previous steps have been successful by running your PCR product on an agarose gel.
- Sequencing the DNA barcode: DNA sequencing is used to read the sequence of As, Ts, Cs, and Gs in the DNA barcode. Typically, you send your PCR products and additional sequencing primers to a company that specializes in Sanger sequencing.
- Bioinformatic analysis: The sequencing company will provide you with DNA sequence data that can be uploaded and analyzed via a service like BLAST, which compares the sequence you provide to a reference database of species-specific sequences to identify a match and calculate the statistical significance of a match.
Easy ways to get started
There are some well-established barcodes you can use to identify a wide variety of species. We’ve listed them below with a few great kits from miniPCR bio to help you get started!
| Type of organism | DNA region | Lab kits and protocols |
| Fungi | ITS | Fungal DNA Barcoding Kit and Fungal sequencing add-on |
| Bacteria | 16S | Bacterial DNA Barcoding: 16S rRNA Amplification |
| Animals | COI | DNA Learning Center: DNA Barcoding 101 |
| Plants | rbcL, matK |
Related resources:
- Mushroom ID Project: Fungal DNA Barcoding Kit
- Bacterial DNA Barcoding Project: 16S rRNA Amplification Kit
- DNA Dots: DNA barcoding explained for students
- International Barcode of Life
- Barcoding with miniPCR in the field
- Blog: Barcoding fungi in your at-home lab!
- Blog: Unlocking fungal secrets with DNA barcoding